Mixture Density Networks
Supervised machine learning models learn the mapping between the input features (x) and the target values (y). The regression models predict continuous output such as house price or stock price whereas classification models predict class/category of a given input for example predicting positive or negative sentiment given a sentence or paragraph. In this notebook, we are going to focus on regression problems and work our way from predicting the target values to learning to approximate the underlying distribution.
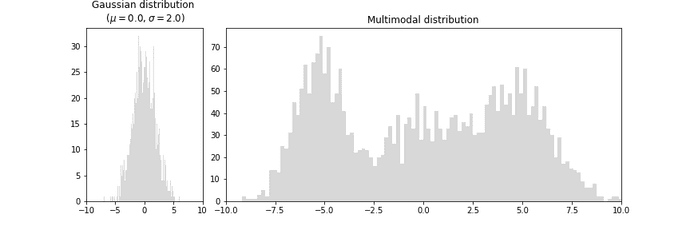
But, why learn distribution when we can directly predict the target values? Well, in most regression problems we assume the distribution of the target value to follow Gaussian distribution (left plot) but in reality, many problems have multiple modalities (right plot) that cannot be solved by directly predicting target values.
We explore this using an example presented in PRML book, where we show that a 2 layer neural network is able to approximate the target values for a given input, but fails when we invert the problem. i.e input becomes the target and vice-versa.
# Add required imports
import numpy as np
import matplotlib.pyplot as plt
plt.style.use('default')
%matplotlib inline
Toy problem
# Example presented in the PRML book
def create_book_example(n=1000):
# sample uniformly over the interval (0,1)
X = np.random.uniform(0., 1., (n,1)).astype(np.float32)
# target values
y = X + 0.3 * np.sin(2 * np.pi * X) + np.random.uniform(-0.1, 0.1, size=(n,1)).astype(np.float32)
# test data
x_test = np.linspace(0, 1, n).reshape(-1, 1).astype(np.float32)
return X, y, x_test
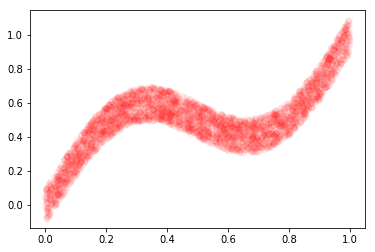
# Plot data (x and y)
X, y, x_test = create_book_example(n=4000)
plt.plot(X, y, 'ro', alpha=0.04)
plt.show()
Neural Network
We will use Tensorflow to build a 2-Layer neural network with fully connected layers to learn the mapping between X and y.
# Load Tensorflow
import os
os.environ['CUDA_VISIBLE_DEVICES'] = '0'
import tensorflow as tf
from copy import deepcopy
# Print tensorflow version
tf.__version__
‘2.0.0-dev20190208’
# Is GPU Available
tf.test.is_gpu_available()
True
# Are we executing eagerly
tf.executing_eagerly()
True
# Build model
def get_model(h=16, lr=0.001):
input = tf.keras.layers.Input(shape=(1,))
x = tf.keras.layers.Dense(h, activation='tanh')(input)
x = tf.keras.layers.Dense(1, activation=None)(x)
model = tf.keras.models.Model(input, x)
# Use Adam optimizer
model.compile(optimizer=tf.keras.optimizers.Adam(lr=lr), loss='mse', metrics=['acc'])
# model.compile(loss='mean_squared_error', optimizer='sgd')
return model
We could play around with model configuration to see how it impact the predictions. I am using the one described in the PRML book (page 274).
# Load and train the network
model = get_model(h=50)
epochs=1000
# Change verbosity (e.g verbose=1) to view the training progress
history = model.fit(X, y, epochs=epochs, verbose=0)
print('Final loss: {}'.format(history.history['loss'][-1]))
# Plot the loss history
plt.plot(range(epochs), history.history['loss'])
plt.xlabel('Epochs')
plt.ylabel('Loss')
plt.title('Training')
plt.show()
Final loss: 0.0034893921427428722
With our model trained and ready for prediction, we test it on simulated data
# Predict the target values for test data and
# plot along with training data
y_test = model.predict(x_test)
plt.plot(X, y, 'ro', alpha=0.05, label='train')
plt.plot(x_test, y_test, 'bo', alpha=0.3, label='test')
plt.legend(bbox_to_anchor=(1.05, 1), loc=2, borderaxespad=0.)
plt.title('Plot train and test data')
plt.show()
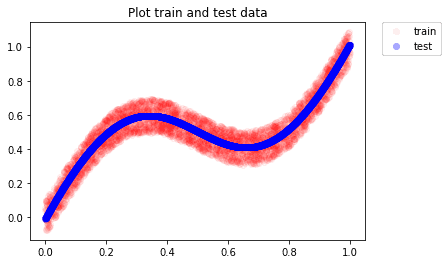
As we can see, the neural network is able to approximate well and we see the predictions following the mean of true target values
Inverse problem
Next, we invert the problem to investigate whether the same/similar model is able to approximate the target values.
# print data shape
print(X.shape, y.shape)
# Deepcopy is not required in this case
# (its more of a habit for me)
flipped_x = deepcopy(y)
flipped_y = deepcopy(X)
# load and train the model
model = get_model(lr=0.09)
# Train the model for large number of epochs
epochs=1500
history = model.fit(flipped_x, flipped_y, epochs=epochs, verbose=0)
print(history.history['loss'][-1])
plt.plot(range(epochs), history.history['loss'])
plt.xlabel('Epochs')
plt.ylabel('Loss')
plt.title('Training')
plt.show()
(4000, 1) (4000, 1)
0.05178604498505592
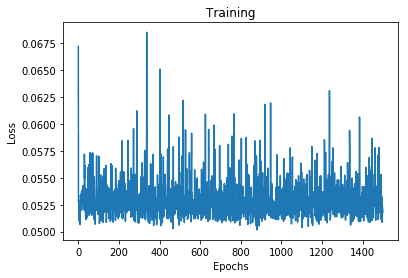
Well, the loss didn’t change much after the first few epochs, let see how well does the model perform
# epochs 500
x_test_inv = np.linspace(0., 1., 4000).reshape(-1, 1)
y_test_inv = model.predict(x_test_inv)
plt.plot(y, X, 'ro', alpha=0.05)
plt.plot(x_test_inv, y_test_inv, 'b.', alpha=0.3)
plt.show()
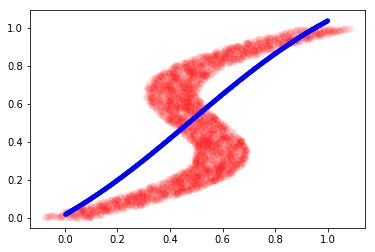
This is the best our model could do because it was confused by the non-gaussian distribution of target values. Note that there are multiple modes in the plot above.
The model could only learn to generate a linear mapping between our inverse data. This motivates the idea of learning the distribution instead of directly mapping the input and output values. The Mixture density network (MDN) does this by learning K different Gaussian parameters for every input data. We see how to implement it the next section
Motivation
<detour>
My motivation to learn MDN comes primarily from the world models paper by Ha and Schmidhuber. The world models paper presents a technique to train an agent, in a reinforcement learning environment, in an unsupervised manner. It makes use of three components: Variational Autoencoder to convert high dimensional space into the low dimension, MDN-RNN to compress temporal respentation and a linear model to determine what action to take to maximize cumulative reward. I intend to cover this in detail in a future post or notebook.
</detour>
MDN
In MDNs, instead of modeling the input (x) -> target (y) mapping by explicitly generating the output values, we learn the probability distribution of each target and sample the predicted output from that distribution. The distribution itself is represented by several gaussians (i.e a mixture of gaussians). A mixture of gaussians is able to represent a complex distribution as shown in the plot titled ‘Multimodal distribution’ at the begining of this post.
For every input x, we learn the distribution parameters namely mean, variance and mixing coefficient. In terms of total output values, there are (l+2)k values where k is the number of gaussians and l is the number of input features. The breakdown of output values is shown below:
- Mixing coefficient : K
- Variance () : K
- Mean() : L * K
MDN provides a generic framework for modeling conditional probability distribution using a linear combination of mixing coefficient and respective component densities:
Neural network
We use a similar neural network as used in previous examples but instead learn the mixing coefficients and component density parameters.
# In our toy example, we have single input feature
l = 1
# Number of gaussians to represent the multimodal distribution
k = 26
# Network
input = tf.keras.Input(shape=(l,))
layer = tf.keras.layers.Dense(50, activation='tanh', name='baselayer')(input)
mu = tf.keras.layers.Dense((l * k), activation=None, name='mean_layer')(layer)
# variance (should be greater than 0 so we exponentiate it)
var_layer = tf.keras.layers.Dense(k, activation=None, name='dense_var_layer')(layer)
var = tf.keras.layers.Lambda(lambda x: tf.math.exp(x), output_shape=(k,), name='variance_layer')(var_layer)
# mixing coefficient should sum to 1.0
pi = tf.keras.layers.Dense(k, activation='softmax', name='pi_layer')(layer)
Display model summary for inspection.
model = tf.keras.models.Model(input, [pi, mu, var])
optimizer = tf.keras.optimizers.Adam()
model.summary()
Model: "model_11"
__________________________________________________________________________________________________
Layer (type) Output Shape Param # Connected to
==================================================================================================
input_12 (InputLayer) [(None, 1)] 0
__________________________________________________________________________________________________
baselayer (Dense) (None, 50) 100 input_12[0][0]
__________________________________________________________________________________________________
dense_var_layer (Dense) (None, 26) 1326 baselayer[0][0]
__________________________________________________________________________________________________
pi_layer (Dense) (None, 26) 1326 baselayer[0][0]
__________________________________________________________________________________________________
mean_layer (Dense) (None, 26) 1326 baselayer[0][0]
__________________________________________________________________________________________________
variance_layer (Lambda) (None, 26) 0 dense_var_layer[0][0]
==================================================================================================
Total params: 4,078
Trainable params: 4,078
Non-trainable params: 0
__________________________________________________________________________________________________
Loss
The loss function with respect to component weights w is given by
The component density is given by
Always handy to keep the latex in front during implementation.
Although the following two functions now look reasonably intuitive, it took me several hours to get it right. Without the eager mode, it would have taken me a lot longer to debug and identify the issue.
# Take a note how easy it is to write the loss function in
# new tensorflow eager mode (debugging the function becomes intuitive too)
def calc_pdf(y, mu, var):
"""Calculate component density"""
value = tf.subtract(y, mu)**2
value = (1/tf.math.sqrt(2 * np.pi * var)) * tf.math.exp((-1/(2*var)) * value)
return value
def mdn_loss(y_true, pi, mu, var):
"""MDN Loss Function
The eager mode in tensorflow 2.0 makes is extremely easy to write
functions like these. It feels a lot more pythonic to me.
"""
out = calc_pdf(y_true, mu, var)
# multiply with each pi and sum it
out = tf.multiply(out, pi)
out = tf.reduce_sum(out, 1, keepdims=True)
out = -tf.math.log(out + 1e-10)
return tf.reduce_mean(out)
Lets verify whether our implementation is correct by comparing the values with numpy version
# calc_pdf(3.0, 0.0, 1.0).numpy()
calc_pdf(np.array([3.0]), np.array([0.0, 0.1, 0.2]), np.array([1.0, 2.2, 3.3])).numpy()
array([0.00443185, 0.03977444, 0.06695205])
# Numpy version
def pdf_np(y, mu, var):
n = np.exp((-(y-mu)**2)/(2*var))
d = np.sqrt(2 * np.pi * var)
return n/d
print('Numpy version: ')
pdf_np(3.0, np.array([0.0, 0.1, 0.2]), np.array([1.0, 2.2, 3.3]))
Numpy version: array([0.00443185, 0.03977444, 0.06695205])
We also verify that MDN loss function works as expected
loss_value = mdn_loss(
np.array([3.0, 1.1]).reshape(2,-1).astype('float64'),
np.array([[1.0, 0.0, 0.0], [1.0, 0.0, 0.0]]).reshape(2,-1).astype('float64'),
np.array([[0.0, 0.1, 0.2], [0.0, 0.1, 0.2]]).reshape(2,-1).astype('float64'),
np.array([[1.0, 2.2, 3.3], [1.0, 2.2, 3.3]]).reshape(2,-1).astype('float64')
).numpy()
assert np.isclose(loss_value, 3.4714, atol=1e-5), 'MDN loss incorrect'
Train
While the current dataset is small enough to be used directly, here we use the dataset API because it is trivial to load data from numpy using dataset API.
# Use Dataset API to load numpy data (load, shuffle, set batch size)
N = flipped_x.shape[0]
dataset = tf.data.Dataset \
.from_tensor_slices((flipped_x, flipped_y)) \
.shuffle(N).batch(N)
We will use the tf.function decorator to convert python code into tensorflow graph code (the goodness of keras API and eager execution makes tensorflow 2.0 a lot more fun)
@tf.function
def train_step(model, optimizer, train_x, train_y):
# GradientTape: Trace operations to compute gradients
with tf.GradientTape() as tape:
pi_, mu_, var_ = model(train_x, training=True)
# calculate loss
loss = mdn_loss(train_y, pi_, mu_, var_)
# compute and apply gradients
gradients = tape.gradient(loss, model.trainable_variables)
optimizer.apply_gradients(zip(gradients, model.trainable_variables))
return loss
Define the model and main training loop.
losses = []
EPOCHS = 6000
print_every = int(0.1 * EPOCHS)
# Define model and optimizer
model = tf.keras.models.Model(input, [pi, mu, var])
optimizer = tf.keras.optimizers.Adam()
# Start training
print('Print every {} epochs'.format(print_every))
for i in range(EPOCHS):
for train_x, train_y in dataset:
loss = train_step(model, optimizer, train_x, train_y)
losses.append(loss)
if i % print_every == 0:
print('Epoch {}/{}: loss {}'.format(i, EPOCHS, losses[-1]))
print every 600 epochs
Epoch 0/6000: loss 1.0795997381210327
Epoch 600/6000: loss -0.8137094378471375
Epoch 1200/6000: loss -0.9803442358970642
Epoch 1800/6000: loss -1.0087451934814453
Epoch 2400/6000: loss -1.0302231311798096
Epoch 3000/6000: loss -1.0441244840621948
Epoch 3600/6000: loss -1.051422357559204
Epoch 4200/6000: loss -1.0547912120819092
Epoch 4800/6000: loss -1.0570876598358154
Epoch 5400/6000: loss -1.0589677095413208
# Let's plot the training loss
plt.plot(range(len(losses)), losses)
plt.xlabel('Epochs')
plt.ylabel('Loss')
plt.title('Training loss')
plt.show()
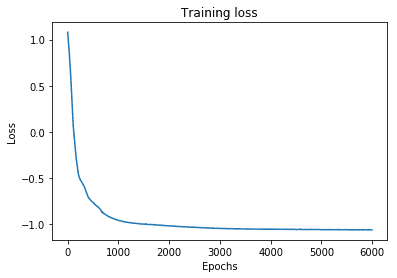
Next, we predict the mixing coefficients and the component density parameters () for test data and visualize the approximate conditional mode.
def approx_conditional_mode(pi, var, mu):
"""Approx conditional mode
Because the conditional mode for MDN does not have simple analytical
solution, an alternative is to take mean of most probable component
at each value of x (PRML, page 277)
"""
n, k = pi.shape
out = np.zeros((n, l))
# Get the index of max pi value for each row
max_component = tf.argmax(pi, axis=1)
for i in range(n):
# The mean value for this index will be used
mc = max_component[i].numpy()
for j in range(l):
out[i, j] = mu[i, mc*(l+j)]
return out
# Get predictions
pi_vals, mu_vals, var_vals = model.predict(x_test)
pi_vals.shape, mu_vals.shape, var_vals.shape
# Get mean of max(mixing coefficient) of each row
preds = approx_conditional_mode(pi_vals, var_vals, mu_vals)
# Plot along with training data
fig = plt.figure(figsize=(8, 8))
plt.plot(flipped_x, flipped_y, 'ro')
plt.plot(x_test, preds, 'g.')
# plt.plot(flipped_x, preds2, 'b.')
plt.show()
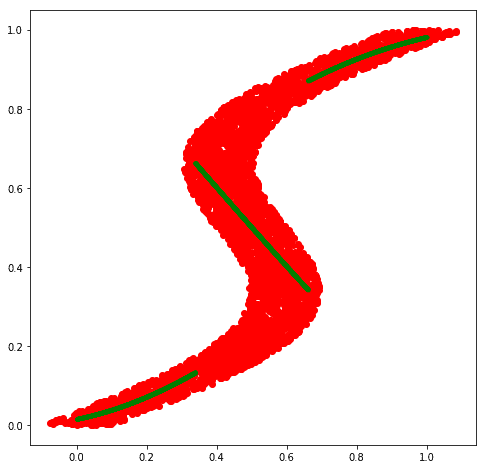
The mean of most probable density (approx conditional mode) looks promising as it does seem to capture different modalities present in the target values.
# Display all mean values
# fig = plt.figure(figsize=(8, 8))
# plt.plot(flipped_x, flipped_y, 'ro')
# plt.plot(x_test, mu_vals, 'g.', alpha=0.1)
# plt.show()
Now that we have learned the distribution for each input value, we can sample a number of points (e.g 10) from the distribution and generate a dense set of predictions instead of picking just one.
def sample_predictions(pi_vals, mu_vals, var_vals, samples=10):
n, k = pi_vals.shape
# print('shape: ', n, k, l)
# place holder to store the y value for each sample of each row
out = np.zeros((n, samples, l))
for i in range(n):
for j in range(samples):
# for each sample, use pi/probs to sample the index
# that will be used to pick up the mu and var values
idx = np.random.choice(range(k), p=pi_vals[i])
for li in range(l):
# Draw random sample from gaussian distribution
out[i,j,li] = np.random.normal(mu_vals[i, idx*(li+l)], np.sqrt(var_vals[i, idx]))
return out
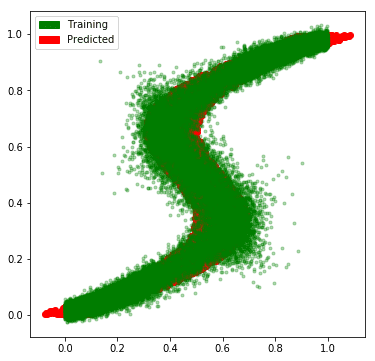
sampled_predictions = sample_predictions(pi_vals, mu_vals, var_vals, 10)
And, we see that our network has learned the approximate the multimodal distribution rather well.
# Plot the predictions along with the flipped data
import matplotlib.patches as mpatches
fig = plt.figure(figsize=(6, 6))
plt.plot(flipped_x, flipped_y, 'ro', label='train')
for i in range(sampled_predictions.shape[1]):
plt.plot(x_test, sampled_predictions[:, i], 'g.', alpha=0.3, label='predicted')
patches = [
mpatches.Patch(color='green', label='Training'),
mpatches.Patch(color='red', label='Predicted')
]
plt.legend(handles=patches)
plt.show()
Conclusion
In this post, we saw how multi modal regression problems can be solved using mixture density networks. We also saw that imperative programming environment in tensorflow 2.0 provides a pythonic way of writing code and autograph decorator allows us to convert the python code into the performant tensorflow graph.
Tensorflow 2.0, exciting times ahead.